Summary of Results
Parameters Inferred
We focused on estimating the following ecological parameters:
| Parameter | True Value | Predicted Value |
|---|---|---|
individuals_local | 7,469 | 6,745 |
individuals_meta | 14,455 | 11,844 |
species_meta | 131 | 142 |
speciation_local | 4.36×10⁻⁵ | 5.08×10⁻⁵ |
mutation_rate | 5.79×10⁻⁶ | 5.20×10⁻⁶ |
These results show that the neural estimator can recover parameters with reasonable accuracy, even in high-dimensional, stochastic systems.
Model Acceleration
To scale training, we utilized CUDA-enabled GPUs, which allowed:
- 10,000+ simulations to be processed in milliseconds
- Efficient posterior sampling using PyTorch + Masked Autoregressive Flows (MAF)
- Rapid iteration and evaluation over large prior spaces
Visual Insights
- Species Richness Dynamics: Our SBI approach successfully captured patterns in species richness over time.
- Posterior Distributions: Density plots confirmed that the model learned well-separated, biologically meaningful parameter spaces.
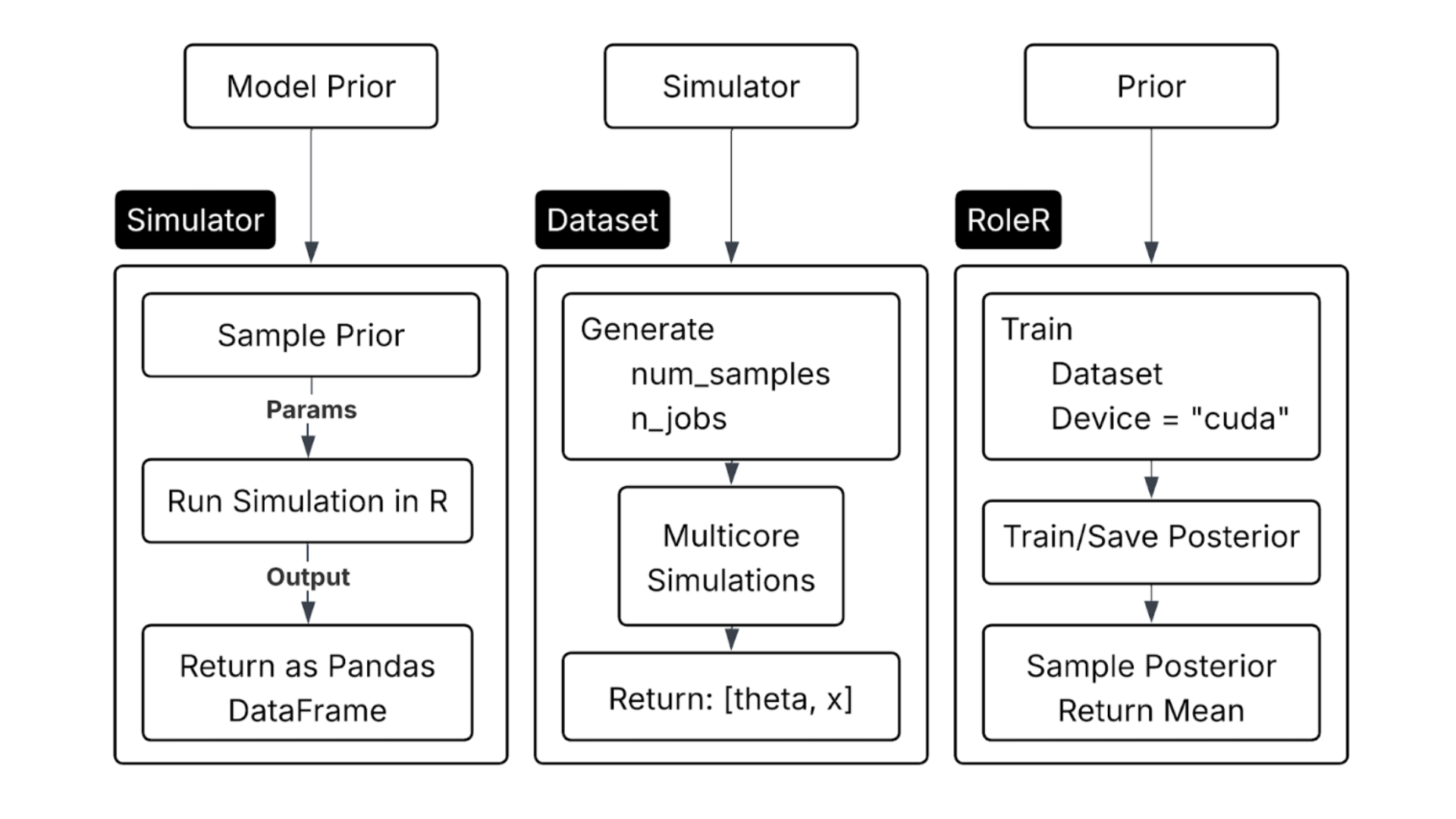
Chart 1: Simulation dynamics of species richness across time.
Chart 2: Posterior distributions inferred for selected parameters.
Implications
This approach demonstrates that Likelihood-Free Inference (LFI), powered by deep generative models, can bridge the gap between complex ecological simulations and real-world data. It enables scalable, interpretable inference without requiring explicit likelihood functions.
As biodiversity datasets continue to grow, these tools provide essential infrastructure for modern ecological research.
